Compound-Specific Stable Isotope Methodology

A central focus of research in the Ocean Ecogeochemistry Lab is the development of compound-specific stable isotope analysis (CSIA) tools and knowledge that push the analytical frontier in food web ecology. At its core, this work seeks to quantify the magnitude, direction, and causes of differential isotopic fractionation of carbon, nitrogen, and hydrogen during the synthesis and subsequent trophic modification of individual compounds, such as amino acids and fatty acids. In doing so, this work advances the development of cutting-edge new CSIA tools while also improving our understanding of organismal nutritional ecology.
Through controlled laboratory experiments, our lab seeks to better understand the underlying biochemical and physiological mechanisms behind how organisms acquire, modify, and allocate dietary resources. This approach has provided groundbreaking advances in how ecologists and oceanographers study resource utilization and food web dynamics in complex ecosystems. The two primary foci of our controlled laboratory studies are to 1) develop, expand, and refine amino acid carbon isotope fingerprinting to reconstruct the phylogenetic identity of primary producers fueling food web dynamics, and 2) quantify mechanisms of nitrogen isotope fractionation associated with trophic transfer.
Amino Acid Fingerprinting:
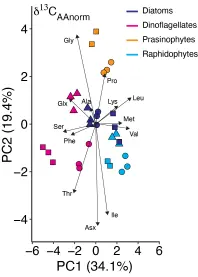
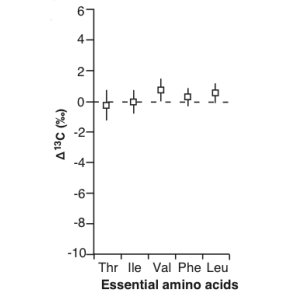
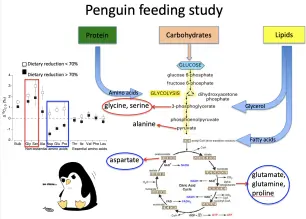
Advances in compound-specific stable isotope analysis have greatly enhanced studies of carbon flow in ocean environments by analyzing δ13C values of specific biochemical compounds, particularly amino acids (AAs). There is tremendous metabolic diversity in the synthesis pathways and associated isotope effects for creating individual amino acids among diverse primary producers. This is particularly true for essential amino acids, which often require seven or more individual enzymatic steps to synthesize. This diversity results in unique stable isotope “fingerprints” of the primary producers that made those amino acids. These unique essential amino acid carbon isotope fingerprints of primary producers propagate through food webs virtually unmodified. This is because while primary producers and bacteria can synthesize essential amino acids de novo, most animals do not possess the necessary enzymatic pathways to synthesize these amino acids at a rate sufficient for normal growth and therefore must incorporate essential amino acids directly from their diet. This direct incorporation of essential amino acids results in little to no change in stable isotope value during a trophic transfer from diet to consumer.
Early efforts to develop amino acid fingerprints focused on widely divergent taxonomic groups of primary producers, e.g., terrestrial plants, aquatic macroalgae, and fungi. Through culture work and targeted wild harvest, we are exploring the specificity of amino acid carbon isotope fingerprints to accurately distinguish among marine-relevant primary producers (e.g., diverse micro- and macroalgae that fuel many marine food webs). Our lab is also examining the stability of amino acid fingerprints across variations in oceanographic and growing condition. This work involves growing primary producers under varying conditions and examining how their amino acid carbon isotope fingerprints change in response. We are also working on developing statistical normalization procedures to account for variations in environmental condition. This work will allow us to compare fingerprints across space and time where growing conditions are not conserved. Additionally, we are working on experiments to examine the influences of nutritional quality and gut microbes on the synthesis and propagation of primary producer fingerprints through food webs.
Trophic discrimination factors:


The concept of trophic position provides a valuable architectural framework for characterizing consumer-resource relationships within food webs. The bulk stable isotope approach to reconstructing trophic dynamics has become widely applied in food web ecology. However, consumer bulk stable isotope values reflect both the stable isotope value of the base of the food web and the number of trophic transfers and associated degree of trophic modification that nitrogen has undergone from the base to the consumer. As a result, it can be very challenging to determine whether differences in consumer bulk δ15N values reflect variability in the δ15Nbaseline, trophic positions, Δ15NC-D values, or some combination of all of these factors. The CSIA of amino acid nitrogen has the potential to disentangle the relative influences of baseline and trophic variability on consumer stable nitrogen isotope value. The power of this approach lies in the differential 15N enrichment of individual amino acids during trophic transfer.
We now commonly divide protein amino acids into two basic groups, termed the “trophic” and the “source” amino acids. The trophic amino acids undergo significant isotopic fractionation during transamination and deamination, owing to their close linkage to the rapidly cycling internal glutamate pool. In contrast, the source amino acids show relatively little trophic fractionation between δ15Nbaseline and measured values in upper trophic level consumers. The ability to independently estimate the δ15Nbaseline from source amino acids and the number of trophic transfers from trophic amino acids has now provided ecologists with a powerful tool to calculate consumer TPCSIA that is internally indexed to the δ15Nbaseline. Our research uses controlled feeding experiments to examine the magnitude, direction, and underlying causes of amino acid stable nitrogen isotope fractionation during trophic transfer. We are particularly interested in understanding the causes of variation in trophic amino acid nitrogen isotope fractionation. Recent work has suggested that trophic amino acids show highly variable trophic fractionation as a function of diet quality and consumer mode of nitrogen excretion. We are also exploring the roles of microbes in altering source amino acid δ15N values in upper trophic level consumers.
Non-essential amino acids and diet quality:
There is growing interest in the potential that carbon isotopes of non-essential amino acids can play in learning about organismal growth, physiology, nutritional condition, and disease. Nonessential AAs can either be de novo biosynthesized from a bulk carbon pool or directly routed from dietary protein, with typically highly variable trophic discrimination factors values across taxa and diet types. Our lab has been exploring how changes in non-essential amino acid carbon isotope values can record information about the macromolecular sources of diet and diet quality (e.g., protein vs lipids), measures of nutritional stress, and metrics of exogenous vs endogenous feeding. This work often takes the form of controlled feeding experiments where we modify things like diet type and quality, growing condition, and physiological conditions studies to observe how non-essential amino acid carbon isotope dynamics respond. We’ve also worked with wild caught species in well constrained systems to examine natural variation in non-essential amino acid isotope dynamics. For instance, we've been working to develop quantitative metrics of how physiologically stressful events like reproduction, molting, and migration impact non-essential amino acid carbon isotope dynamics in penguins, whales, and seals in the wild. This sort of work will open new doors for exploring organismal health, growth, and reproductive capacity in the future.
Publications: (# indicates student author)
Ohkouchi N, Chikaraishi Y, Close HG, Fry B, Larsen T, Madigan DJ, McCarthy MD, McMahon KW, Nagata T, Naito YI, Ogawa NO, Popp BN, Steffan S, Takano Y, Tayasu I, Wyatt ASJ, Yamaguchi YT, Yokoyama Y (2017) Advances in the application of amino acid nitrogen isotopic analysis in ecological and biogeochemical studies. Organic Geochemistry DOI: 10.1016/j.orggeochem.2017.07.009 (authors in alphabetical order after first)
McMahon KW, McCarthy MD (2016) Embracing variability in amino acid δ15N fractionation: Mechanisms, implications, and applications for trophic ecology. Ecosphere 7:e01511 (Special issue on Biomarkers in Trophic Ecology)